February 15, 2023
New preprint alert from our lab!
“Improving Molecular Machine Learning Through Adaptive Subsampling with Active Learning”
https://doi.org/10.26434/chemrxiv-2023-h8905
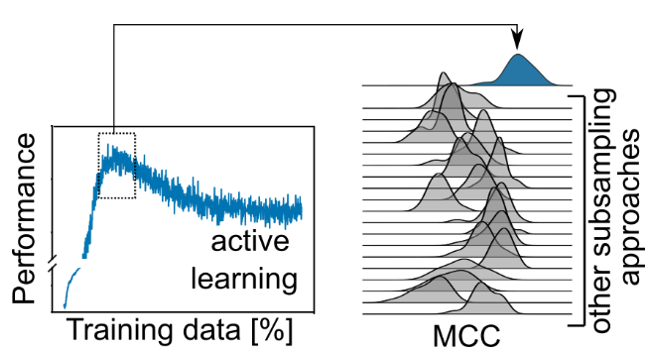
We and others had previously shown that active learning can learn on a fraction of the training data and still achieve competitive performance (see http://dx.doi.org/10.1016/j.ddtec.2020.06.001 ). We were wondering whether there are cases where active learning outperforms training on the full data.
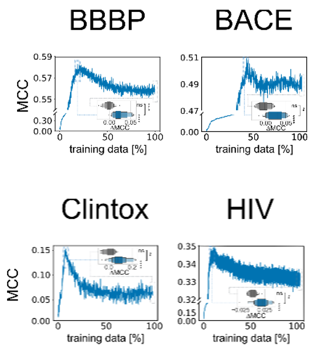
When we used scaffold-splitting on all MoleculeNet benchmark tasks, we observed that active learning performance on the test set would peak and then decrease until a model trained on the full data performed significantly worse.
It is known that active learning often selects a more balanced dataset compared to the underlying data distribution – and that is certainly true here as well. But that does not seem sufficient to explain all the benefits since simple balanced subsampling does not lead to the same performance improvements.
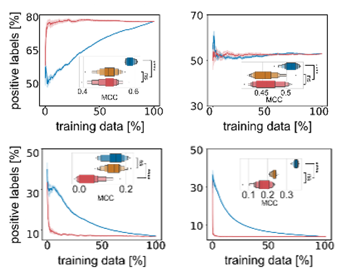
Instead, it looks like active learning consistently selects the same datapoints for training even if it is initialized with different initial data. Active learning selects a core set of most valuable training data, possibly at the decision boundary of chemical space.
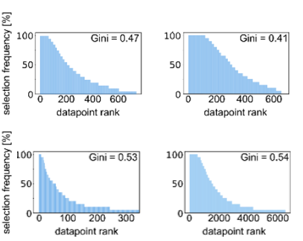
Importantly, active learning is more robust to errors in the training data. Randomly flipping labels in the training data decreases performance of models but active learning can train more robust models even if 20-30% of the data are incorrect.
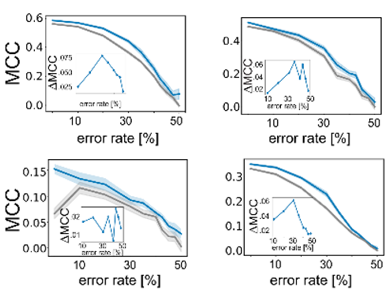
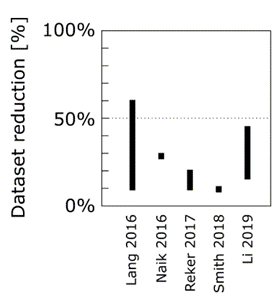
This motivated us to compare the performance of active learning on erroneous data to other established data sampling strategies. Gratifyingly, active learning is never outperformed by any of the other state-of-the-art methods.

Active learning is a powerful approach, and here we showcase its potential to serve as an autonomous data curation method. Code to apply this method to your data is on GitHub and includes a notebook that runs subsampling out of the box.
https://github.com/RekerLab/active-subsampling
We are intrigued about what we learned about active learning – incl. robustness to erroneous data and overfitting on constraint search spaces. Stay tuned for more retrospective and prospective active learning work coming out of our laboratory.