Active machine learning
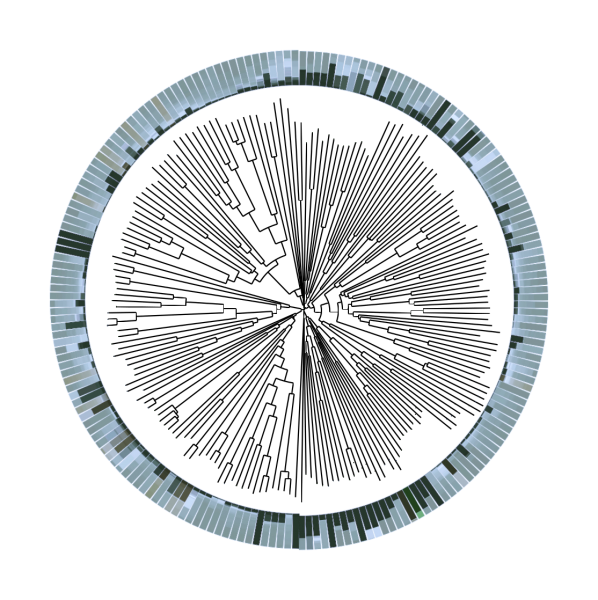
Machine learning is an invaluable tool to detect patterns in big data and streamline experiments by prioritizing the most promising candidates, as indicated by the increasing number of companies in this space. However, machine learning performance is tightly coupled to the size and quality of the available data. Major efforts in machine learning research aim at improving predictive architectures to deal with complex, sparse, and noisy data. We take an alternative approach and instead use machine learning in the generation of high quality data. To this end, we let the machine learning model decide which data it would most profit from. Coupled to laboratory automation, such active machine learning workflows can drive experimentation and autonomous optimization. Multiple of our studies have shown that active learning can improve machine learning performance and identify promising new solutions rapidly, for example in the context of drug discovery, chemogenomics, and chemical reaction condition optimization. However, multiple practical challenges remain. Currently, we are investigating what the most efficient learning strategies are, how groups of experiments should be selected, how to select training data, the effects of data augmentation, and how to integrate orthogonal data. We tailor active machine learning platforms and investigate model interpretability to guide biochemical experiments or complex computations and apply the resulting models in the analysis and development of therapeutics.
Predicting properties of therapeutics
We harness big and small biochemical and clinical data to predict unknown properties of approved or investigational therapeutics. Such efforts help explain mechanisms of action, guide drug-repurposing efforts, predict drug-drug interactions, direct potency optimizations, and identify side-effects of drugs. The latter is particularly relevant for mechanisms that are historically underrecognized, such as interference with epigenetic targets. When analyzing approved therapeutics, we also have to consider their "inactive ingredients", which can potentially lead to allergic reactions in hypersensitive patients or unknowingly change drug exposure. These underappreciated effects directly impact patient outcomes and guide future drug discovery and development efforts. Through validating such predictions in vitro or using clinical data, we can improve our understanding of currently available therapeutics, guide the design of novel therapeutic candidates, as well as gain actionable insights for personalized medicine.
Designing new formulations
Coupling our predictive models and assay technologies enables us to efficiently design novel drug delivery solutions. These technologies can boost the efficacy and safety of medications, for example by enabling better targeting, modulating the metabolism of drugs, or optimizing ADMET profiles. Through the application of machine learning, we can hasten the discovery of such materials. By specifically focusing on re-purposing food and drug additives, we generate drug delivery solutions with accelerated pathway for potential approval. For example, we have identified food additives that can slow down drug metabolism, invented coatings for enzymatically-controlled drug release, and designed novel and tunable nanoparticles from small molecular excipients that increase their anti-cancer effectiveness in vivo. Currently on-going efforts investigate the in vivo potential of functional excipients that reduce the side-effects and toxicity of the delivered drug.